June research news roundup
UW–Madison research helps launch first FDA-cleared blood test for Alzheimer’s disease
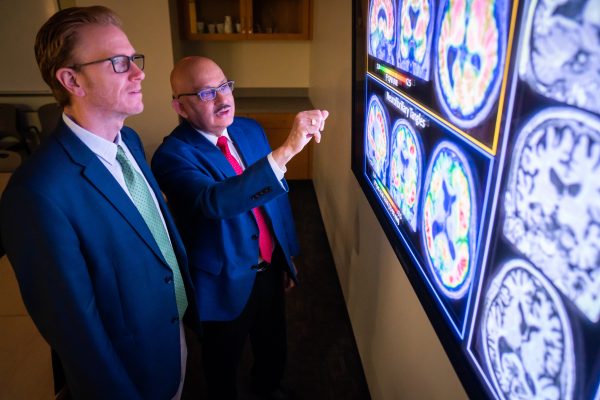
In a crucial advancement for Alzheimer’s disease diagnosis, the U.S. Food and Drug Administration (FDA) granted clearance for an Alzheimer’s disease blood test on May 16, 2025. The test is an in vitro diagnostic (IVD) assay capable of detecting amyloid — a key protein involved in Alzheimer’s disease — in blood. Data samples used to evaluate the validity of the test are from research conducted at the University of Wisconsin–Madison.
The Lumipulse G pTau217/ß-Amyloid 1-42 Plasma Ratio is for the early detection of amyloid plaques associated with Alzheimer’s disease in adult patients, aged 55 years and older, exhibiting signs and symptoms.
The Wisconsin Alzheimer’s Disease Research Center (ADRC) and the Wisconsin Registry for Alzheimer’s Prevention (WRAP) provided 40% of the samples used in the validation study to evaluate the plasma p-tau217/Aβ42 ratio as a reliable biomarker of amyloid pathology in the brain. At the UW–Madison Alzheimer’s disease programs’ biofluids laboratory, these biomarker levels were measured in cerebrospinal fluid (CSF); and then demonstrated that levels of these proteins were congruent in blood plasma samples from the same individuals.
“This is spectacular news and a breakthrough in identifying reliable blood biomarkers for Alzheimer’s disease,” said Sanjay Asthana, MD, founding director of the Wisconsin ADRC. “Our ADRC and WRAP programs were instrumental in this achievement, underscoring the strength and vision of our biomarker research initiatives.”
Sterling Johnson, PhD, principal investigator of the WRAP study, said the FDA approval will have clinical and wide significance. “This milestone transforms years of patient-centered research into a tool physicians can now use to benefit their patients and guide care,” he said. “As one of the early leaders in Alzheimer’s biomarker research, the UW Alzheimer’s program is at the forefront of this progress.”
Read the full story here.
UW–Madison researchers find hidden genetic clues upping cardiovascular disease risk
Researchers at the University of Wisconsin–Madison have uncovered new evidence in a decades-old genetic mystery, discovering how a group of genetic variations in a long-mysterious region of the human genome can put people at higher risk of cardiovascular disease.
Thanks to past studies of the whole human genome that drew associations between particular variations and disease, scientists have known for nearly 20 years that alterations in a section of human chromosome 9 called the 9p21.3 locus contribute to increased risk of developing coronary artery disease, CAD. The specific variations make atherosclerosis, a dangerous thickening and stiffening of the coronary arteries with plaque, more likely.
However, it was not well understood how the variations in this region lead to an increased risk for CAD. To solve that mystery, UW–Madison scientists coaxed induced pluripotent stem cells, which are living donor cells reprogrammed into a blank-slate state, int o developing into the smooth muscle cells found in the walls of blood vessels.
o developing into the smooth muscle cells found in the walls of blood vessels.
Valentina Lo Sardo
In the lab of UW–Madison stem cell scientist Valentina Lo Sardo, genetics graduate student Elsa Salido found that smooth muscle cells carrying the risky version of 9p21.3 behaved differently than typical cells in healthy blood vessels, instead developing some of the features of cells seen in cartilage and bone.
Read the full story here.
Valentina Lo Sardo (above), lead researcher on the project and a professor of Cellular and Regenerative Biology in the UW School of Medicine and Public Health and member of UW–Madison’s Stem Cell and Regenerative Medicine Center.
Deep learning method identifies transition states in protein conformational changes
In a study published in Nature Communications, researchers at the University of Wisconsin–Madison introduced a deep learning method capable of automatically identifying transition states in protein conformational changes, a key process that underpins many biological functions.
This new tool promises to accelerate the study of biomolecular dynamics and could have wide-reaching applications in drug design, biomolecular engineering, and materials science.
This study is a collaborative effort between Prof. Xuhui Huang’s group (Department of Chemistry) and Prof. Sharon Li’s group (Department of Computer Sciences) at the University of Wisconsin–Madison.
Read the full story here.
Game Incubator Event Fuels Educator, Research Partnership at UW–Madison
Researchers from UW–Madison recently teamed with state educators and game designers in a day-long competition called Game Incubator to pitch ideas for a new, free educational learning game that could be played by hundreds of thousands of young students across Wisconsin and beyond. Field Day Lab hosted the event, which was part of GameWorks, a UW–Madison School of Education initiative launched in March in a partnership between the lab, the Wisconsin Center for Education Research (WCER) and the Wisconsin Department of Public Instruction (DPI).
The brainstorming and design event will produce a game that supports student learning about life sciences, such as bioengineering, genetics, conservation biology, human geography, or fungal biology, by letting players assume the role of a field researcher at work.
“We are trying to draw inspiration from the real, lived experiences of these researchers, the work they do and the problems they encounter,” said Jim Mathews, education director of UW–Madison’s Field Day Lab, which will spend $250,000 in grant funds to build out one game idea. The winning project will be selected in late summer. “We want to translate the authentic research process into a game that fits a classroom.”
Read the full story here: https://www.wcer.wisc.edu/news/detail/game-incubator-event-fuels-educator-research-partnership-at-uwmadison